Conventional wisdom says “superbugs” emerge in places like India or Africa and move to the US through global travel. That was certainly the case with the Ebola virus recently and the NDM-1 bacteria from India.
However, an interesting research letter in Nature Genetics examined the global evolution and spread of a particularly nasty epidemic caused by Clostridium difficile. Last year, the CDC listed C. difficile as one of the top 3 antimicrobial resistance threats in the US. This disease already kills more than 14,000 Americans a year and sickens a quarter million more, a bigger toll by far than the global death count from Ebola.
But before Gov. Christie tries to close the borders, this paper tells us that epidemic strains of C. difficile first emerged in North America, especially in the northeast US.
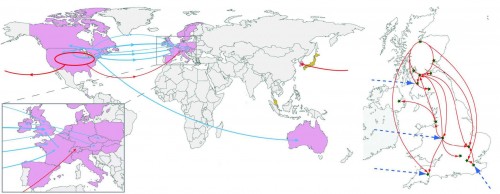
The study comes from the Wellcome Trust Sanger Institute in Cambridge, England, applying whole genomic sequencing to 151 clinical samples collected around the world over the past 25 years. Here’s the map, showing the emergence of two distinct fluoroquinolone-resistant (FQR) epidemic strains from the US (red) and the US/Canada (blue):
This highly infectious disease originated in North America and spread to health care facilities across the globe. Note also the subsequent transmission within the UK (box on right, red lines) after the 4 initial “invasions” from the US (directly and via France).
Why did FQR C. difficile emerge in the US? It is probably our own fault, for overusing antibiotics:
Furthermore, our data suggest that the acquisition of resistance to commonly used antibiotics is a major feature of the continued evolution and persistence of C. difficile 027/BI/NAP1 in healthcare settings. It is notable that fluoroquinolone antibiotics were one of the most commonly prescribed antibiotic classes in North America during the late 1990s and early 2000s, such that it is during this time that selective pressure for the acquisition and maintenance of fluoroquinolone resistance within healthcare settings would have been at its highest, explaining the near simultaneous emergence of more than one clone of FQR C. difficile 027/BI/NAP1.
A second important finding related to agriculture, food and the environment:
The derived phylogeny suggests that C. difficile 027/BI/NAP1 has transmitted between human and non-human sources in both directions. For example, multiple isolates from food sources or animals in Arizona were derived from a historical Arizona human isolate (BI-2 from Tucson, 1991). In a sub-lineage of FQR1, a number of isolates from food sources or animals were found in the exact same position in the phylogeny as human isolates, suggesting an identical genotype; the tip of this sub-lineage being a human isolate from New Jersey (BI-13). These data suggests C. difficile 027/BI/NAP1 transmits through the food chain, and human C. difficile could contaminate the environment. However, a more comprehensive strain collection would be needed to confirm this. (emphasis added)
Re-read that last sentence: we really don’t know the scope of transmission because of the small number of samples in this study (n=151). The recently-announced US National Strategy to Combat Antibiotic-Resistant Bacteria calls for much greater investment in “One Health” surveillance so we can better understand what we are up against. Considering the threat, this should be a major priority.
@koutterson