While antibiotics degrade with use through resistance, most casual observers assume that vaccines are immune to these effects. They would be wrong. Resistance is simply evolutionary change under environmental pressure. For influenza vaccines, evolutionary changes render prior flu vaccines relatively ineffective each year, which is why influenza samples are collected in Southeast Asia each year and shared with 5 national labs to help develop the new vaccine each year. (In the US, the final decision is made by the VRBPAC – the Vaccine and Related Biological Products Advisory Committee at the FDA).
Beyond influenza, we have mounting evidence that some vaccines affect the ecological balance of the susceptible and non-susceptible serotypes of the relevant pathogens. For example, the HPV-4 vaccine targets the 4 most serious serotypes of the HPV virus, but many dozens of other HPV serotypes are not covered by the vaccine. Likewise, the original pneumococcal conjugate vaccine (PCV-7) targeted 7 serotypes of pneumococcus; the newer one targets 13, but coverage remains incomplete.
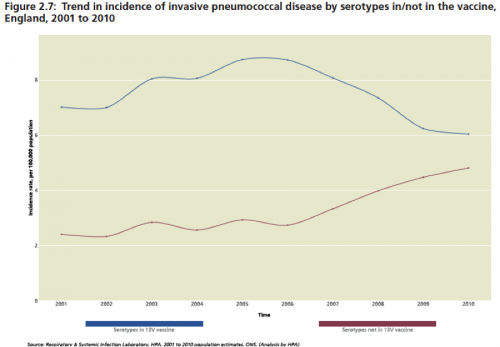
Over time, we are seeing evidence of “serotype replacement,” in pneumococcus and HPV meaning that as the vaccines work to remove the targeted types from the human environment, the non-targeted serotypes expand to fill the ecological niche. In addition, and even more troubling, is when vaccine-targeted pneumococcus acquire elements by recombinations, including capsule switches that render them protected from the vaccine (Science). Some data on the effect of PCV-13:
Annual Report of the Chief Medical Officer, Vol. 2, 2011; see also LID 2011;11(10):760-68.
Which brings me to the paper Marc Lipsitch & colleagues just published in Philosophical Transactions B. Using stochastic math models, they explored how antibiotic use interacted with serotype replacement in pneumococcus. The presence of antibiotics confers an advantage on resistant strains, which affects the emergence of resistance:
Under neutral conditions no emergence was observed. With a 1% advantage, emergence was observed in a minority of realizations for the high-burden setting. At 3% and above, emergence was nearly universal, indicating that antibiotic use is a key determinant of the emergence of resistant lineages following PCV introduction. (emphasis added)
And from the discussion:
We have integrated these factors into a single model with the conclusion that antibiotic pressure is likely the most important driver of the emergence of resistant, NVT [non-vaccine-type] lineages…(emphasis added)
This combination of observed data and other modelling work lend credence to our finding that the expansion of a previously rare antibiotic-resistant lineage is probable following the introduction of a PCV targeting common resistant types. So long as vaccination targets only a subset of pneumococcal serotypes, antibiotic pressure will likely lead to the emergence of resistant lineages. This may be facilitated in settings that combine high or intermediate drug pressure with high carriage prevalence. (emphasis added)
The solution is not to abandon vaccines, but to develop better vaccines that cover all serotypes and in the meantime to understand the impact on resistance.
And if you have read this far, perhaps you are interested in joining the Presidential Advisory Council on Combating Antibiotic-Resistant Bacteria. Deadline for nominations closes on May 13, 2015, so hurry!
@koutterson